
Dassanayake Lab
COMPARATIVE
FUNCTIONAL
& EVOLUTIONARY
GENOMICS

Data and tools generated in our group
Genome Assembly and Annotation
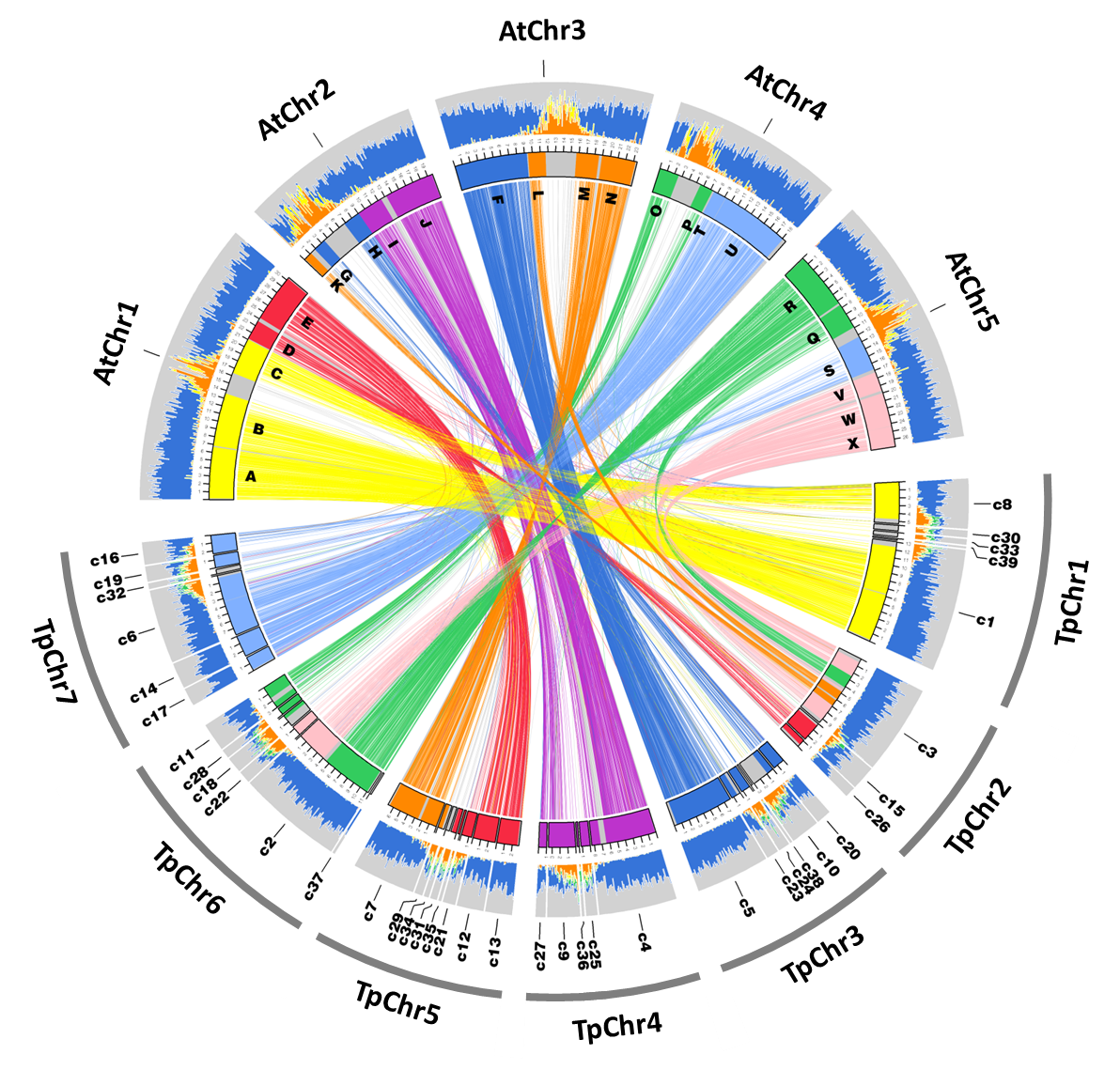
Schrenkiella parvula Version 2.0 (Apr. 2014) (formerly Thellungiella parvula or Eutrema parvula)
Genome assembly: Genome sequence (fasta)
Genome annotation: CDS (fasta),
Notes on the update:
- Genome contig/scaffolds and gene models renamed reflecting the chromosome model.
- Redundant genome contig/scaffolds were removed.
Schrenkiella parvula Version 1.0 (Aug. 2011) (Dassanayake et al., 2011)
Genome assembly: Genome sequence (fasta)
Genome annotation: CDS (fasta)
RNAseq and Transcriptome Data
Cell type-specific responses to salinity - the epidermal bladder cell transcriptome of Mesembryanthemum crystallinum (Oh et al., 2015)
Genome structures and transcriptomes signify niche adaptation for the multiple-ion-tolerant extremophyte Schrenkiella parvula (Oh et al., 2014)
- S. parvula genome version 2.0 (see above) was used for this study.
Comparative genomics tools to identify shared evolutionary genomic variation
CL_Finder
Tool to identify and annotate orthologous genomic units including genes in multiple genomes
OrthNet
Tool to create orthology based networks
Pipeline described at doi: https://doi.org/10.1101/236299 and can be downloaded from GitHub