
Dassanayake Lab
COMPARATIVE
FUNCTIONAL
& EVOLUTIONARY
GENOMICS


Create Novel Genomic and Transcriptomic Resources
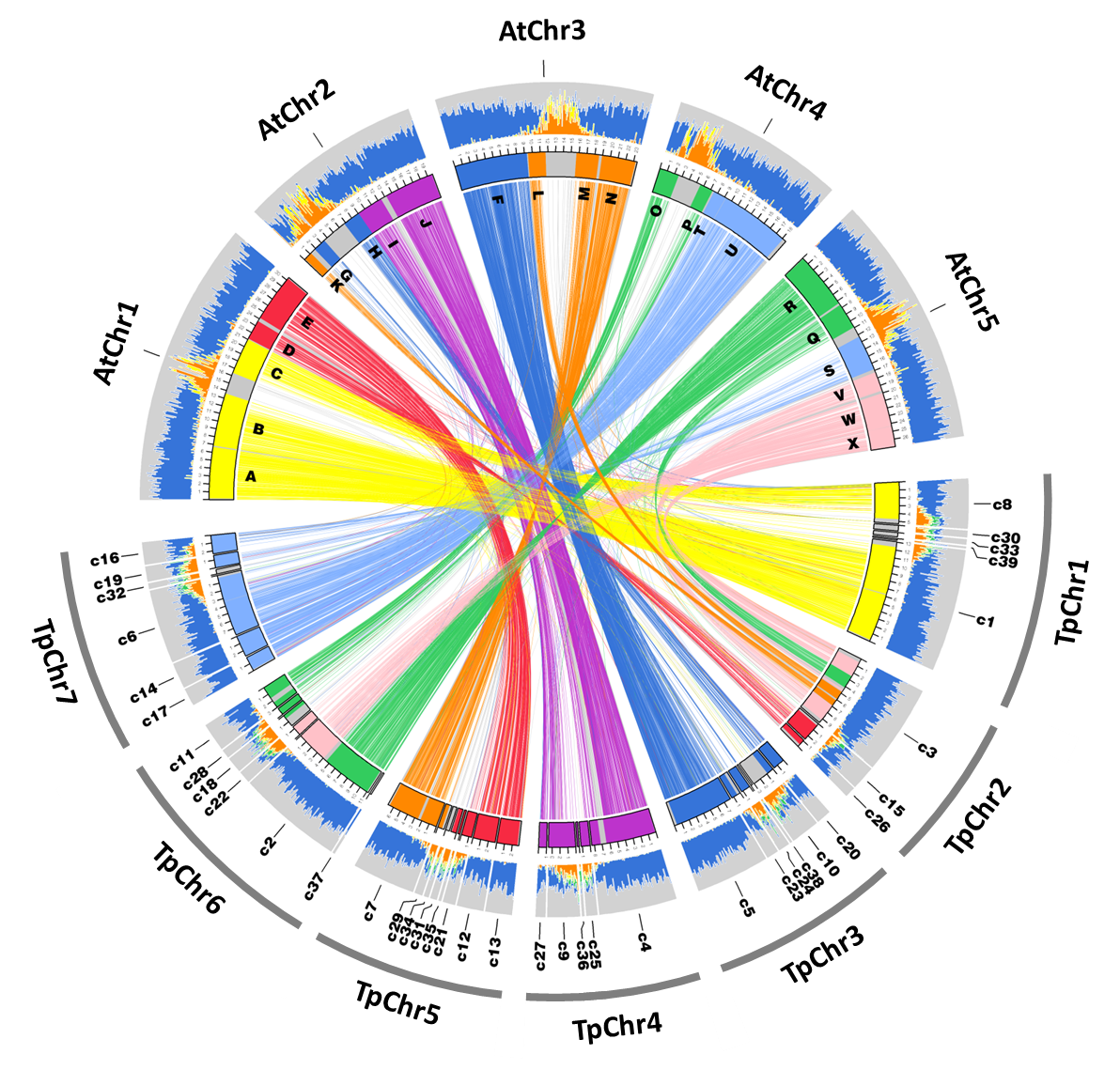
Genomes and transcriptome atlases for extremophyte models


Schrenkiella parvula
gDNA
Adaptations?
LIFESTYLE?
(Dassanayake et al., Nature Genetics, 2011)
Next Generation Sequencing (NGS) technologies have enabled high throughput sequencing of genomes and transriptomes of previously unexplored species. We have used NGS to create reference genomes and transcriptomes of plants adapted to multiple abiotic stresses. We use hybrid assembly pipelines to assemble short reads (< 300 bp) and long-reads (> 100 kbp) into longest possible contiguous sequences that can serve as reference sequences for comparative genomics and transcriptomics studies.
(See the RESOURCES page for updates on the genome assembly and annotations of extremophyte models.)
Assembled genome of Schrenkiella parvula

Rhizophora mangle, Twin Cays, Belize
Image by John Cheeseman



Images by Hyewon Hong